RNA drugs one step closer to be being used in cancer treatment
In recent years, RNA molecules, with the ability to affect or turn off pathogenic genes, have become promising drug candidates in several areas. However, it has been a challenge to develop techniques to deliver the RNA molecules into the cells where they have an effect. Researchers at Lund University in Sweden have now developed a sensitive technique that makes it possible to study the delivery into the cell, and have shown a possible way to effectively deliver RNA drugs to tumours. The study has now been published in Nature Communications.
Published 2020-04-20
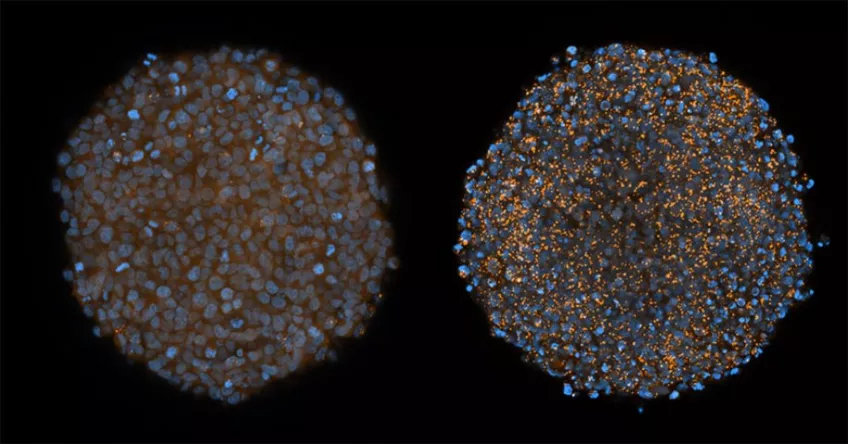
Membrane damage caused by chloroquine in tumour spheroids. Cervical cancer cells cultivated as small tumours in test tubes, so-called tumour spheroids. The spheroid on the right has been treated with the substance chloroquine, which resulted in widespread membrane damage in the cells. This damage is seen as orange spots. The tumour cell nuclei are blue.
DNA and its chemical relative, RNA, are molecules that all living organisms use for storing information and carrying out different functions in the cell. In the study in question, the researchers have studied a type of RNA drug called siRNA. During the 1990s, researchers discovered that siRNA, small double-stranded RNA molecules, could be used to turn off virtually any gene. The phenomenon was named RNA interference. In 2006, the discovery was awarded the Nobel Prize in Physiology or Medicine. There were considerable hopes that RNA interference could be used in the treatment of virus infections, cancer and other diseases.
“Two siRNA drugs have been approved by the FDA, but as yet, no drug has been approved for clinical use in cancer”, says cancer researcher and physician Anders Wittrup at Lund University and Skåne University Hospital who led the study.
One considerable advantage of using RNA molecules is that they can be rapidly developed and produced. The major problem for all sorts of RNA drugs is getting these molecules into the interior of the cell, the so-called cytosol, where they have an effect. The size of the SiRNA molecule – about 50 times larger than a typical drug molecule – is a factor. In the study in question, the researchers studied siRNA molecules linked with cholesterol, which means that they are taken up effectively by most tumour cells.
“Even if we can make tumour cells take up siRNA, it has been observed that 99 per cent appears to get caught in a type of cellular waste dump, the so-called lysosome”, explains Anders Wittrup.
The researchers then examined if, among other things, the use of the malaria drug chloroquine could cause damage in the lysosome, so that the siRNA drug could enter further into the cell. Being able to study this required new imaging techniques in order to see what happens in the cell.
“The completely new microscopy methods make it possible to study in detail when lysosomes and other structures in the cell are opened up for RNA delivery. This is a technique that is in high demand in both academic research and within the pharmaceutical industry”, says doctoral student Hampus Du Rietz.
“We have shown how chloroquine can be used so that the siRNA molecules enter the cytosol, i.e. the space between the cell membrane and the nucleus of the cell”, says Anders Wittrup.
This way of opening the lysosomes, which previously encapsulated the RNA molecules and thus prevented their effect, means that the main obstacle for the use of siRNA and other RNA-based drugs is now potentially on the way to being removed.
“We see a siRNA effect that is 50 times higher in cell culture studies and also effective delivery in the experimental tumours cultivated outside the body, so-called tumour spheroids, that could not previously be reached by siRNA”, says Anders Wittrup.
More studies are needed before RNA drugs are put in clinical use. The research team in Lund has now gone on to studies of more tumour types and other methods of delivering RNA into tumour cells.
“Our findings open up exciting possibilities for how RNA can reach into the interior of cells. I think it will play a crucial role in developing RNA drugs for the treatment of diseases for which we currently lack effective drugs”, concludes Anders Wittrup.
Publication in Nature Communications: Imaging small molecule-induced endosomal escape of siRNA
Contact:
Anders Wittrup, researcher at Lund University, Department of Clinical Sciences, LUCC-Kamprad, Wallenberg Centre for Molecular Medicine and physician at Skåne University Hospital
+46 761 726872
anders [dot] wittrup [at] med [dot] lu [dot] se
The research was funded with support from the Swedish Society for Medical Research (SSMF), the Gunnar Nilsson Cancer Foundation, Mrs Berta Kamprad’s Cancer Foundation, the Märta Winkler Foundation, the Knut and Alice Wallenberg Foundation and Region Skåne.
Researchers behind the publication
Contact:
Anders Wittrup, researcher at Lund University, Department of Clinical Sciences, LUCC-Kamprad, Wallenberg Centre for Molecular Medicine and physician at Skåne University Hospital
+46 761 726872
anders [dot] wittrup [at] med [dot] lu [dot] se (anders[dot]wittrup[at]med[dot]lu[dot]se)
THE CORONAVIRUS CONNECTION
The current coronavirus outbreak has clarified the need for the rapid development and production of new vaccines and drugs, which is one of the main strengths of RNA-based substances.
For example, a number of experimental coronavirus vaccines, so-called mRNA vaccines, are under development. One such mRNA vaccine, developed by the American biotechnology company Moderna, was recently the first coronavirus vaccine to have started clinical trials in human subjects. In addition to this, several pharmaceutical companies have also begun to develop antiviral drugs, to be used for already affected patients, based on siRNA. What these efforts have in common is an attempt to utilise what is theoretically a simpler process for developing RNA-based drugs compared with traditional pharmaceutical substances.